Extensions:2.6/Py/Scripts/Import-Export/PDB
| UI location | File -> Import -> Protein Data Bank (.pdb)
File -> Export -> Protein Data Bank (.pdb) | ||
|---|---|---|---|
| Usage | Import: Choose the pdb file, the options and go onto 'Import'. The atomic structure is then automatically imported afterwards.
Export: Select all objects and structures and export them. | ||
| Version | 1.5 (2012-11-10) | Author(s) | Blendphys (Clemens Barth) |
| Blender | >= 2.64 | License | GPL |
| Category | Import-Export | ||
| Note(s) | Last update of this page: 2018-04-13
2. Other details about the ad-don can be found here: other details 3. Please, use Blender version >= 2.64 | ||
| File name | see files in directory ../scripts/addons/io_mesh_pdb |
|---|---|
| Current version download | io_mesh_pdb.zip |
| External Python Modules or dependencies | No dependencies |
| Data | A custom data file can be used for the colours and radii of the elements. You can download it from here: http://development.root-1.de/X-Download/atom_info.dat |
| Warning | / |
|---|---|
| Links | Atomic Blender personal pages |
| Release Log | Forum |
目次
Quick install
The addon is in the official version of Blender. In the menu choose File, then User Preferences ... and switch to Addons on the top. Search for the Atomic Blender - PDB script and activate it. If the script shall be present after each start of Blender choose Save As Default.
If you want to have the Atomic Blender - Utilities panel, visit this page: Atomic Blender - Utilities panel
Forum
The addon can be discussed in this forum: http://blenderartists.org/forum/showthread.php?235003-Atomic-Blender-for-loading-PDB-files-into-Blender-2.5
PDB files, examples
Play around with these PDB files: PDB files
Examples, recent work
Examples, highlights
This is indeed a highlight, a movie that has been done by the Femto-ST CNRS institute in Besançon (France) (Creator: Sébastien Coget, Researchers: Frank Palmino and Frederic Cherioux)! It demonstrates the combination of the Atomic Blender addons and Blender, and shows that very professional movies can be done for research by Blender. The movie was rendered with cycles. Other movies from the Femto-ST can be found here.
Examples, literature
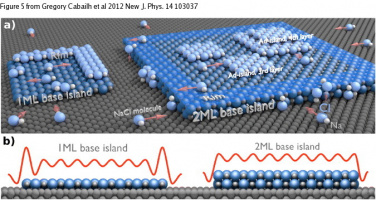
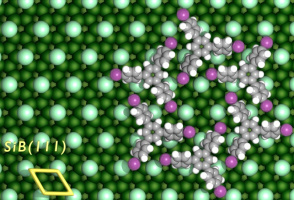
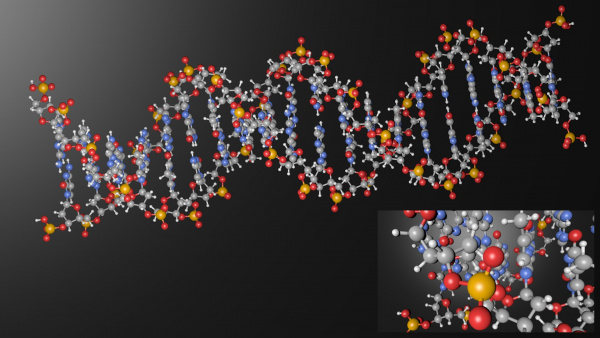
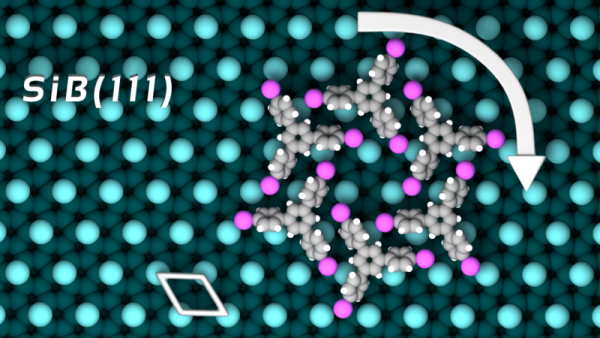
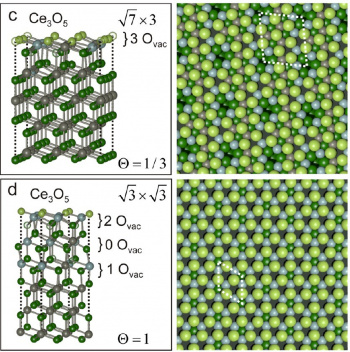
Here are some Cover Pictures and figures, which were produced with Blender and the Atomic Blender addons for high impact journals:
Examples, other
Also quite nice:

What is it good for?
- Producing fancy graphics of structures (atoms, molecules, clusters, surfaces, etc.) for high-impact journals
- 3D scene: Visualisation and analysing structures
- 3D scene: Scientific discussions
- Education
- Having fun
Features
supported
- Atoms (ATOM and HETATM)
- Sticks (CONECT)
- Atomic and van der Waals radii
- Custom data file for radii (pre-defined, atomic and van der Waals) and colours
unsupported:
- No complex structures, only atoms and sticks
Notes
Other details about the addon can be also found here: http://development.root-1.de/Atomic_Blender.php; This page shows some other very nice examples: http://development.root-1.de/Atomic_Blender_PDB_Examples.php
Importing PDB files
Using Atomic Blender is quite easy:
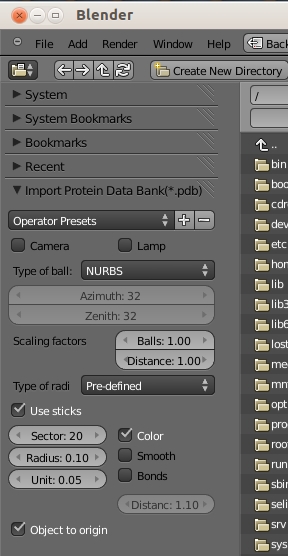
- Got to: File » Import » Protein Data Bank (.pdb) - see Import Protein Data Bank image.
- In the file dialog you find some options on the left. Scroll down to the panel if necessary. Make a choice of options (explained below).
- Search your PDB file and press the button Import Protein Data Bank (*.pdb). Your structure will then be loaded.
All distances and radii are in Angstrom and equal the units of Blender.
Options - file dialog window
Operator Presets - This property is useful to store all the properties you can see below by a key that one can save as follows: First, choose your options by modifying the properties below. Then, go onto + and add a new key and go on okay. - The next time when you want to load a PDB structure, you can choose the key and all options are restored.
Camera and lamp - The camera and lamp are useful for obtaining a quick impression of a loaded structure.
If the button Camera is active, a camera is put into the scene. The camera is 10 times the size of the structure far away from the center of the structure. The camera automatically points to the structure. In the Camera View the camera can be positioned via ⇧ ShiftF for instance (see manual for Blender). A lamp can be loaded, too. The lamp is automatically positioned between structure and camera.
In order to activate the camera, choose the camera A_camera in the Outliner (top right of the Blender Window). Then go to the 3D View and choose in the menu: File » View » Cameras » Set Active Object as Camera.
Type of ball - The type of ball can be chosen, either 'NURBS' (default), 'Mesh' or 'Meta' balls. If 'Mesh' is chosen the number of sectors in azimuth and zenith directions can be chosen as well.
Scalling factors - The radii of all atoms and all distances can be scaled with the two scaling factors.
Type of radii - With this bool enumerator the type of radii can be chosen. Either one uses pre-defined radii, atomic radii or van der Waals radii.The default values for pre-defined radii are the atomic radii.
Use sticks - If this button is active the sticks are drawn, however, only if explicitly mentioned in the PDB file (via record type CONECT). The input 'Sector' can be used to choose the number of sectors for one stick, whereas option 'Smooth' makes the sticks more smooth. 'Unit' (in Angstrom) is the length of a unit of a stick. Such units are aligned, one after the other one, such that the stick is created. This is done due to a specific Blender 'trick' that makes the loading and rendering faster. The smaller the unit is the better is the length precision but the more vertices are drawn (makes loading longer). Input 'Radius' is the radius of a stick in Angstroms. If the Boolean 'Color' is crossed the sticks have the colours of the atoms (half-half). If 'Bonds' is active, double and triple bonds are shown by doubled or tripled sticks, respectively. The float property underneath can be used to choose the distance between the sticks in units stick diameter. If 'bonds' is not active only one single stick is shown.
Object to origin - Sometimes it may happen that structures in PDB files have an offset. If this button is active, the structure is put into the center of the scene.
When the structure is loaded it can be seen in the 3D View. The components of the structure are listed in the Outliner (top right in the Blender window). All atoms of one type form one group.
Dupliverts structures and center atom
When atomic structures are imported via the PDB or XYZ importer the atoms are put into so-called dupliverts structures, somewhat into groups of elements (e.g. all hydrogens into one dupliverts structure). A dupliverts structure of one element (e.g., hydrogens) consists of a mesh of vertices, which are located at the positions of the atoms. The number of vertices equals the number of atoms. Furthermore, a dupliverts structure has a representative ball, the atom itself (e.g., hydrogen). This ball is then duplicated at all vertices such that the atomic structure is build. Single atoms can be deleted or displaced in the Edit Mode of Blender by modifying the position of the vertices. However, they are always a part of the structure and are not independent objects.
After the import of an atomic structure into the 3D scene of Blender, the atoms and sticks are visible but also an atom (ball) and stick (cylinder) in the center of the structure (left). The center ball and cylinder do not make sense, but they are very important: they are the representative objects of the dupliverts structures. They cannot be deleted or made invisble, they are always there. However, when rendering a picture these balls and cylinders disappear (right). So, after all they do not really disturb.
Exporting to PDB
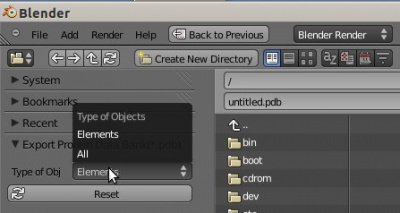
It is possible to export the x, y and z coordinates and names of the atoms or structures (objects) in a Blender scene into a PDB file. Go to: Menu -> File -> Export -> Protein Data Bank (.pdb). The objects can be 'normal' objects or dupliverts structures. They all must be selected and visible in the 3D Blender scene. In the file dialog, one can choose between 'Elements' or 'All'. Option 'Elements' means that only those objects or structures are exported, which contain the name of an element. If option 'All' is chosen, the coordinates of all marked objects and structures are exported. If the name does not contain a name of an element, a '?' is put into the PDB file.
Note: Converting all atoms of a dupliverts structure to real objects
Do the following: Select the whole structure (molecules, surface, or what ever) with the mouse. Go to objects Apply -> Make Dupliverts Real or just push Shift Control A. With this you produce real objects! In the outliner delete the remaining dupliverts, named like 'Carbon', 'Hydrogen', etc.