Extensions:2.6/Py/Scripts/Neuro tool/Presetting Units
Importing meshes from other programs
Importing meshes from TrakEM2 (Fiji)
TrakEM2 is an analysis tool in Fiji (www.ini.uzh.ch/~acardona/trakem2.html), specifically designed for the manual reconstruction and measurement of features in serial microscopy images. The reconstructions can be exported as .obj files.
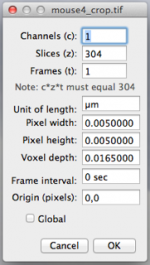
TrakEM2 displays the serial images, and it is important to provide the image dimensions in the Image/Properties panel (see image). These measurements must to be given in microns as these numbers are used by Blender. Therefore, if the voxel size is 5x5x16.5 nm (x,y,z), the parameters are loaded as 0.005x0.005x0165 microns (see image).
TrakEM2 will retain this information, and is assigned to the reconstructed meshes, even when they are exported. Therefore, when importing the mesh to Blender there is no need to rescale the .obj files so a scale factor of 1:1 is given, ie 1 micron corresponding to 1 blender unit.
The setting for the import, therefore, should be to check the box marked 'Use scaling factor', and make sure the scale is 1.00.
Importing meshes from iLastik
iLastik is a open source software created by the group of Fred Hamprecht in Heidelberg (www.ilastik.org), allowing semi-automated segmentation and 3D reconstruction of objects from EM stacks. iLastik does not allow the user to introduce a scale factor to the input stacks. Meshes exported from this software need to be rescaled according to their size directly in Blender.
Consider as an example a cube of serial images representing 5x5x5 microns, corresponding to 1001x1001x304 pixels (x,y,z).
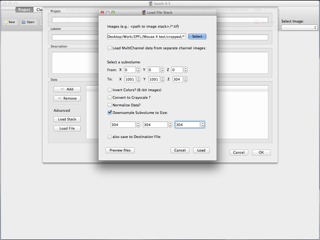
When the .obj files are exported from Ilastik they need to be rescaled upon import to blender. In the case here, the meshes come from a stack whose side length is 5 microns, that comprises 304 pixels. Therefore, in this case the user must check the box labelled 'Rescale by pixel size' and 304 is given as 'Pixel number', and 5 as 'Image side length'. The meshes will then be rescaled correctly once imported.
Important information about using Ilastik on EM image stacks
When the stack is loaded into iLastik, if the voxels are not isotropic (such as in this case), images will need to be resampled to be able to export meshes with the correct x-y-z ratio. Three options are possible:
- oversampling z to the x-y value - downsampling x-y to the z value - oversampling z and downsampling x-y to achieve a reasonable compromise
Oversampling z is not recommended as the smallest misalignment will result in resampled stacks showing artefacts severely affecting the segmentation. The software is high memory demanding. For a MacPro, 8 cores, 16GB RAM, sampling until 450x450x450 pixels represented a reasonable compromise between segmentation accuracy and time consumption to evaluate the segmentation.